IPPCAAS elucidates RNA interference: an ever-advancing cornerstone of plant antiviral defense
Today, the Innovation Team for functional genomics of crop pathogens at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences (IPPCAAS), together with the Laboratory of Virology at Wageningen University & Research (WUR), was invited to publish a review article in Plant Communications (IF = 11.6) entitled “Antiviral RNA interference in plants: increasing complexity and intertwining with other biological processes.” The article systematically summarizes the classical mechanisms of RNA interference (RNAi) in plant antiviral defense, the newly uncovered complex networks, and the prospects for its application in agriculture.
Plant viral diseases are the “invisible killer” in agricultural production, posing a major threat to global food and vegetable security. In the face of complex and variable viruses, plants are not “sitting ducks”; rather, they have evolved a sophisticated immune arsenal in which RNA interference (RNAi) is a central component.
I. RNA interference: the plant’s innate defense system
RNA interference (RNAi) constitutes the first line of defense against viral invasion in plants (Figure 1). Its classical framework comprises two major branches:
PTGS (post-transcriptional gene silencing): After viral RNA enters the cell, Dicer-like proteins (DCLs) cleave viral double-stranded RNA into 21–22 nt virus-derived small interfering RNAs (vsiRNAs). These vsiRNAs are loaded into Argonaute proteins (AGO1/2) to form the RNA-induced silencing complex (RISC), which precisely recognizes and cleaves viral RNAs, thereby blocking their translation and replication.
RdDM (RNA-directed DNA methylation): When viral DNA or double-stranded RNA arising during replication is present, DCL3 processes it into 24-nt vsiRNAs. Through AGO4, these small RNAs recruit DNA methyltransferases to induce methylation of viral DNA, suppressing viral gene transcription.
In addition, RNA-dependent RNA polymerases (RDRs) can use aberrant viral RNAs as templates to synthesize more double-stranded RNA, thereby amplifying siRNA production in a positive-feedback manner. These RNAi signals can also spread via plasmodesmata and move systemically through the phloem, protecting distal, uninfected tissues.
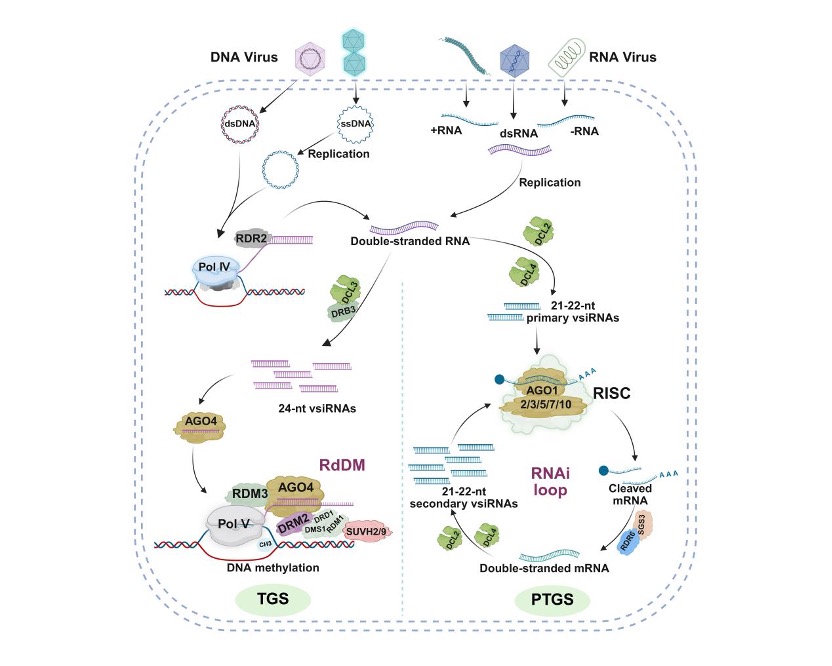
Figure 1. Classical model of antiviral RNAi in plants
II. Beyond the classical framework: non-canonical RNAi pathways and emerging RNA regulatory factors
Antiviral RNAi was once thought to operate in a relatively fixed manner. However, recent studies have revealed non-canonical RNAi pathways in plants. These routes may generate small RNAs of different sizes or participate in antiviral defense in non-traditional ways. Meanwhile, numerous new RNAi regulatory factors continue to be identified and characterized (Figure 2). These findings indicate that RNAi is not a single pathway with a fixed set of effector proteins, but a multilayered, dynamically evolving defense network.
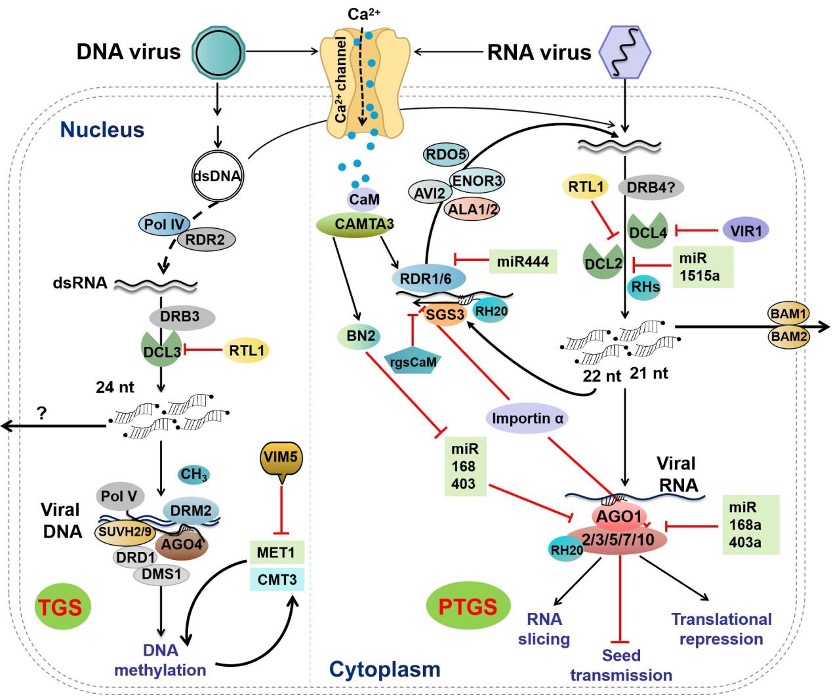
Figure 2. Roles of RNAi pathway components and newly identified regulators in plant antiviral defense
III. Attack–defense dynamics: an “arms race” between host and virus
Viruses do not surrender; they have evolved a variety of RNA-silencing suppressors (RSSs) to counteract RNAi. At the same time, plants continually deploy newly identified positive and negative regulators to modulate RNAi, thereby determining infection outcomes (Figures 2 and 3). Notably, small RNAs may act beyond plants themselves, potentially crossing species boundaries to affect viruses and even insect vectors, forming a tri-partite interaction network among plants, viruses, and insects (Figure 3).
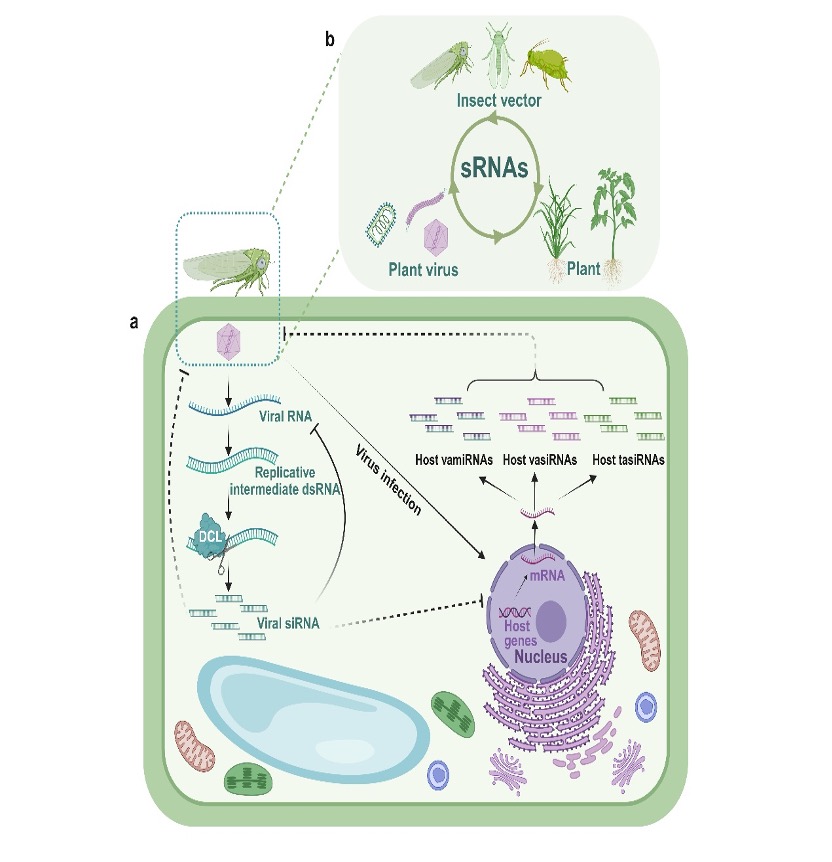
Figure 3. A cross-species model of small-RNA movement among plants, viruses, and insect vectors
IV. Application prospects: from the laboratory to the field
Research on RNAi has not only deepened our understanding of plant immunity, but has also opened new avenues for agricultural application. Beyond conventional transgenic antiviral strategies, exogenous double-stranded RNA (dsRNA) spray technology has attracted wide attention in recent years. Spraying synthetic dsRNA onto crop leaves can activate RNAi and/or pattern-triggered immunity (PTI), achieving “green and environmentally friendly” crop protection. Nevertheless, key questions remain unresolved, including how exogenous dsRNA enters plant cells and is subsequently processed and utilized.
RNA interference is not only an important mechanism by which plants resist viruses; it is also deeply intertwined with hormone signaling, stress adaptation, and even cross-species interactions. With the discovery of more non-canonical RNAi pathways and new regulatory factors, our understanding of this “critical weapon” is steadily expanding. In the future, RNAi is expected to provide more efficient and precise solutions for disease-resistant breeding and sustainable agriculture.
Professor Li Fangfang (IPPCAAS) and postdoctoral researcher Li Xue are co-first authors. Professor Li Fangfang (IPPCAAS), Prof. Richard Kormelink (Wageningen University & Research), and Prof. Zhou Xueping (IPPCAAS/Zhejiang University) are co-corresponding authors. This work was supported by the National Natural Science Foundation of China.
-
 IPPCAAS Advances Joint Prevention and Control of Transboundary Crop Pests and Diseases in Southwest China — Field Demonstration on Monitoring and Control Technologies Held in Jiangcheng, Yunnan
IPPCAAS Advances Joint Prevention and Control of Transboundary Crop Pests and Diseases in Southwest China — Field Demonstration on Monitoring and Control Technologies Held in Jiangcheng, Yunnan -
 IPPCAAS Expert Wins the Society for Invertebrate Pathology Early Career Award
IPPCAAS Expert Wins the Society for Invertebrate Pathology Early Career Award -
 Workshop on Green Control Technology for Corn Pests and Diseases in the Lancang-Mekong Region successfully held in Kunming
Workshop on Green Control Technology for Corn Pests and Diseases in the Lancang-Mekong Region successfully held in Kunming -
 IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety
IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety
