IPPCAAS Achieves Efficient Pyrimidine Base Editing in Rice Using DNA Glycosylases
Recently, the Ecological Safety Evaluation and Utilization Innovation Team for Pest-Resistant Crops at the Institute of Plant Protection, Chinese Academy of Agricultural Sciences (IPPCAAS), published a research paper titled Developing glycosylase-based T-to-G and C-to-K base editors in rice in the renowned plant science journal Plant Biotechnology Journal . The study developed thymine and cytosine base editors (TGBEs and CKBEs, K=G/T) mediated by DNA glycosylases in rice, not only enriching the rice gene-editing toolkit but also providing novel technical means for the directed evolution of endogenous resistance genes in plants, further expanding the application potential of gene editing in crop resistance breeding.
Nucleotide variation is the primary form of genetic variation and plays a critical role in shaping crop genetic diversity and improving agronomic traits. In recent years, DNA deaminase-mediated base editing technologies have been widely adopted in basic research and biological breeding. Cytosine and adenine base editors enable precise C>T and A>G substitutions, correcting defective resistance genes and creating novel superior resistance alleles, accelerating genetic improvement and germplasm innovation for crop resistance. Additionally, guanine base editors based on the guanine DNA glycosylase hMPGv6 have successfully achieved G>T editing in rice. However, direct editing of thymine (T) remains a technical challenge.
In this study, researchers systematically evaluated the base-editing activities of multiple engineered thymine DNA glycosylase variants (TDG-EKΔ, TDG3Δ, and gTBEv3) and cytosine DNA glycosylase variants (CDG4Δ and gCBEv2) in rice by fusing these enzymes with SpCas9n. Results demonstrated that all thymine DNA glycosylases successfully achieved T>G substitutions, with gTBEv3 exhibiting the highest editing efficiency (12.50%–62.50%). CDG4Δ and gCBEv2 also enabled efficient C editing, predominantly generating C>G and C>T substitutions. Beyond base substitutions, TGBEs, and CKBEs induced various nucleotide insertions and deletions (Indels). The DNA glycosylase-based thymine base-editing technology fills the last gap in developing all four base-editing systems in plants. Combined with previously developed cytosine, adenine, and guanine base-editing technologies in rice, the team has now established a comprehensive rice base-editing system, offering robust technical support for the precise improvement and directed evolution of endogenous crop resistance genes, thereby accelerating resistance breeding and germplasm innovation.
Postdoctoral researcher Yongjie Kuang and Ph.D. candidate Xuemei Wu from IPPCAAS are co-first authors of the paper. Associate Researcher Bin Ren is the corresponding author. Researchers Huanbin Zhou, Xueping Zhou, and Associate Researcher Fang Yan from CAAS, along with Professor Dongfang Ma from Yangtze University, also contributed to the study. The research was supported by the National Major Agricultural Biological Breeding Project, the National Natural Science Foundation of China, the Agricultural Science and Technology Innovation Program of CAAS, and the China Postdoctoral Science Foundation.
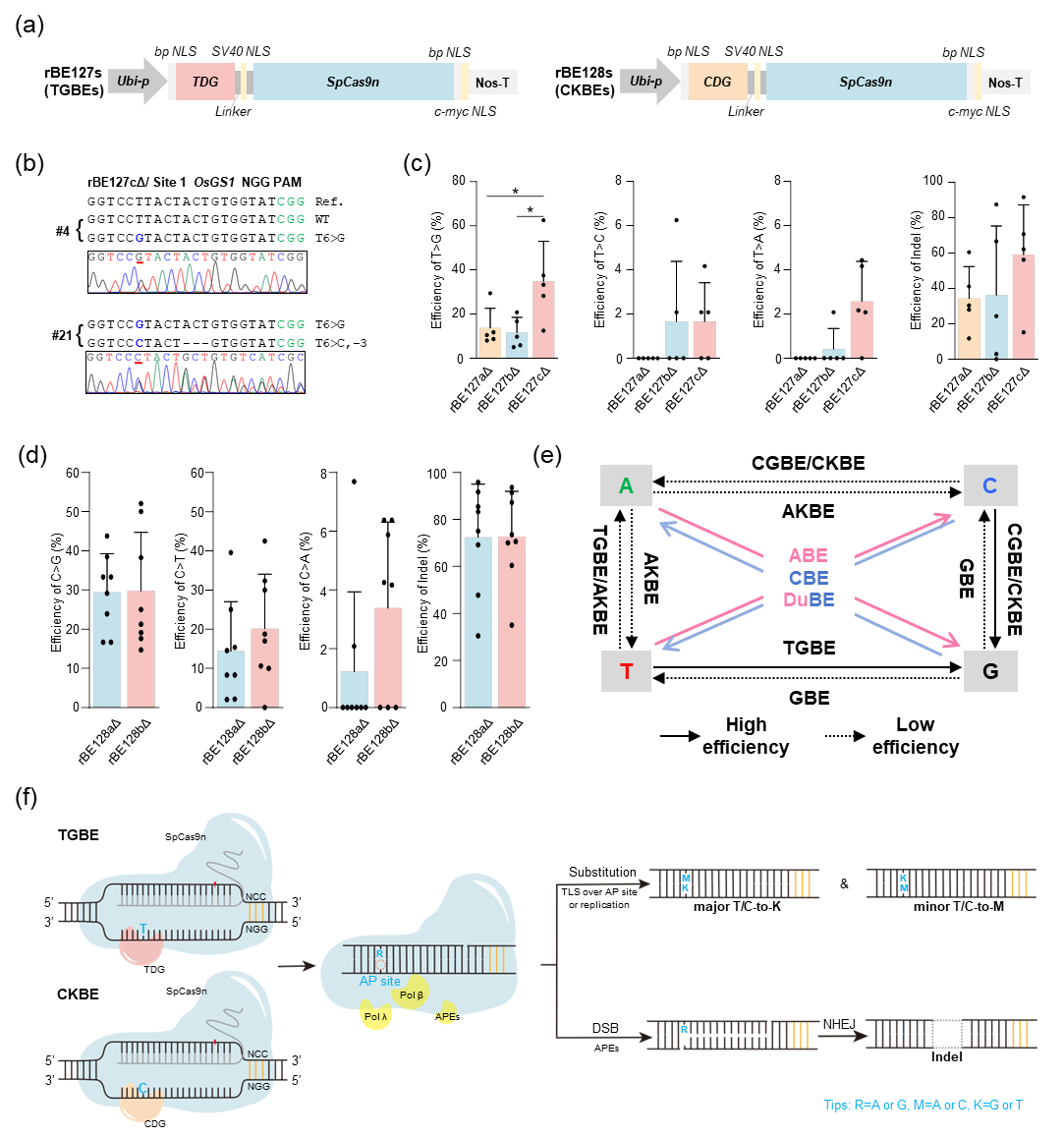
Figure 1: DNA glycosylase-mediated thymine and cytosine base-editing technology in rice.
Source: https://doi.org/10.1111/pbi.70063
-
 IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety
IPPCAAS Experts Visit Australia to Promote In-Depth China–Australia Cooperation in Plant Biosafety -
 China-France cooperation in plant protection was further strengthened
China-France cooperation in plant protection was further strengthened -
 New progress was made for the Lao PDR – China Joint Laboratory for Plant Protection
New progress was made for the Lao PDR – China Joint Laboratory for Plant Protection -
 Collaboration on sustainable agriculture practices highlighted at the MARA China – CABI Joint Lab meeting
Collaboration on sustainable agriculture practices highlighted at the MARA China – CABI Joint Lab meeting
